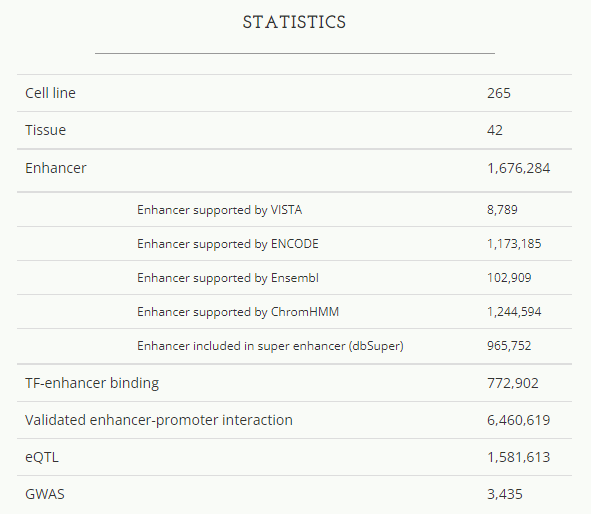
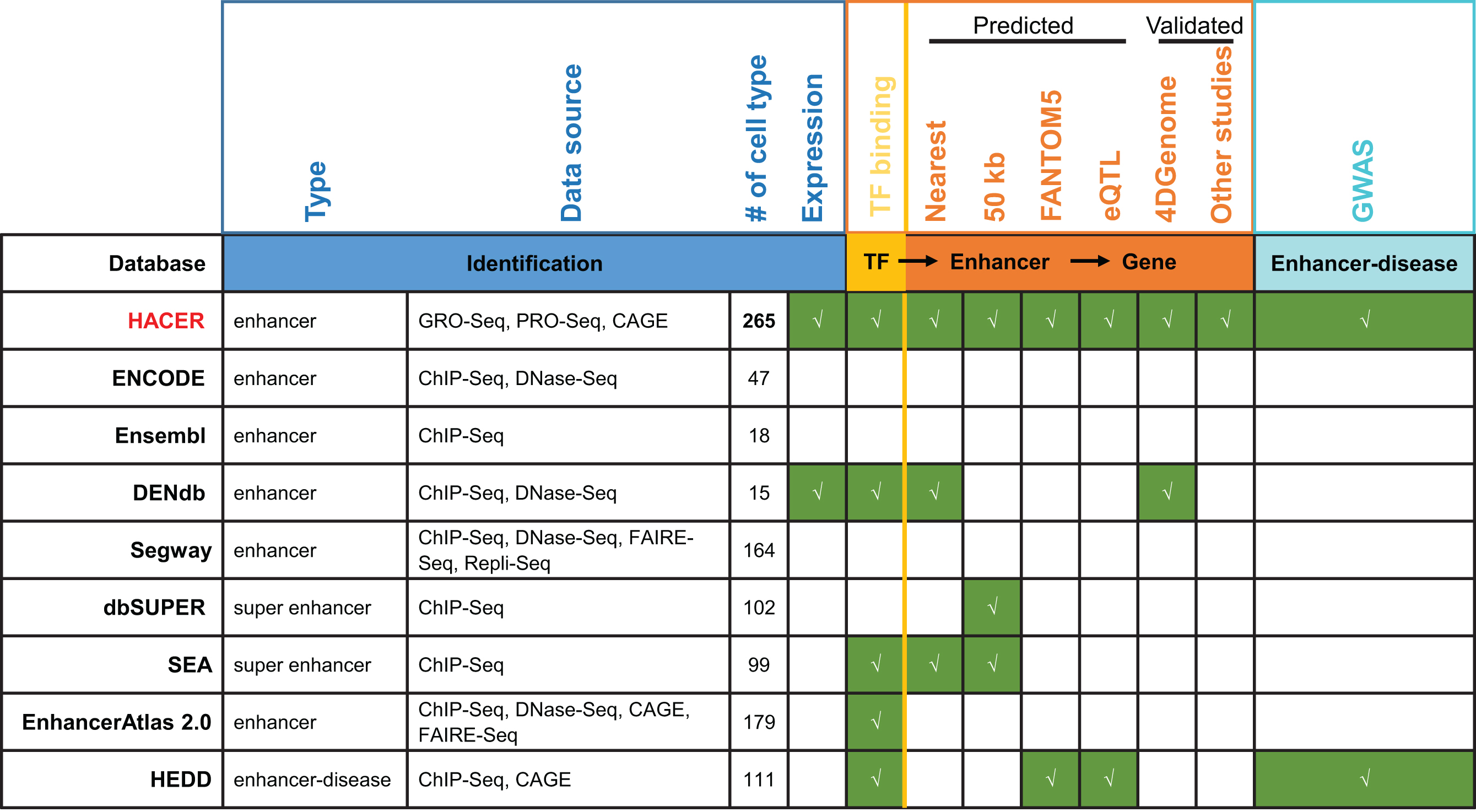
Hacer Enhancer数据库来源
一、VISTA数据库来源的enhancers
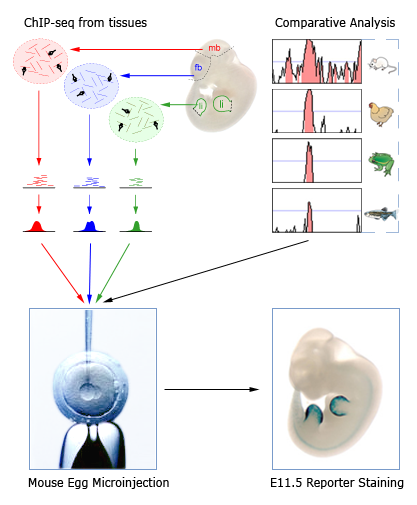
该数据库收录的enhancer元件都是对脊椎动物保守的非编码序列片段和具有潜在enhancer标记的表观遗传数据(比如ChIP-seq)进行的体内增强子效应实验数据。该数据库测试了总共3233个潜在enhancer元件,其中1654个具有enhancer活性。其中,鉴定出的人类活性增强子元件998个。
二、ENCODE来源的enhancers
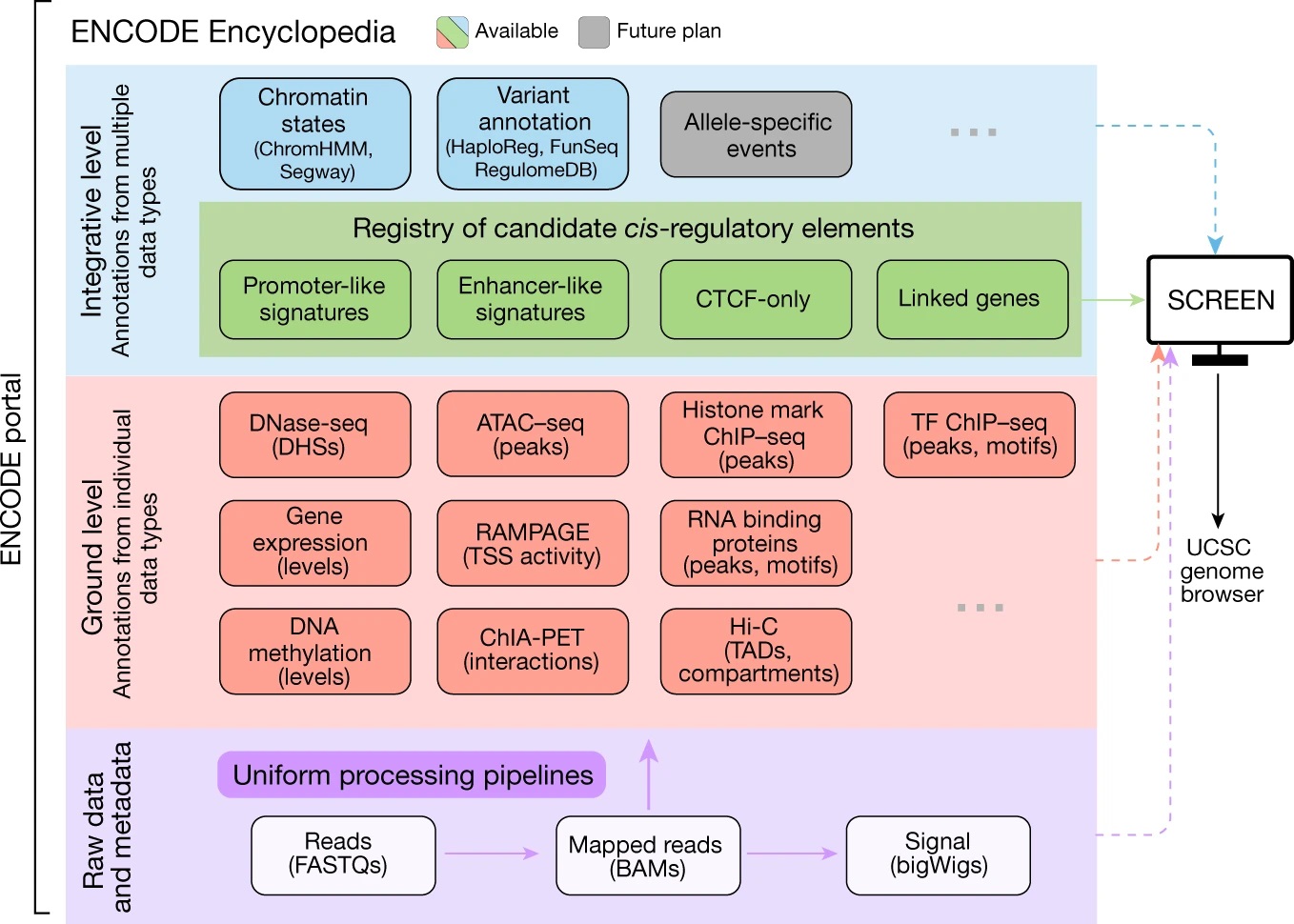
ENCODE enhancer注释的主要思路是结合GENCODE TSS位置信息和增强子相关组蛋白修饰信息,对染色质开放区(DNase-seq或ATAC-seq)进行增强子注释。以TSS peak为核心的高位置特异性的ENCODE开放染色质区域的功能注释(相对TSS的距离,位置可标记为≤200 bp for TSS-overlapping, 200–2,000 bp for TSS-proximal, and >2,000 bp for TSS-distal),将这些区域归为三个主要类:(i) active and poised enhancer-like elements (proximal and distal, 15.3% and 72.1% of human cCREs); (ii) active promoter-like elements (3.7% of human cCREs); and (iii) CTCF-only elements (6.1% of human cCREs),以及一个次要类DNase and H3K4me3 (DNase–H3K4me3; 2.8% of human cCREs)。关于调控元件的注释以及活性状态由DNase、H3K4me3、H3K27ac 或 CTCF这四种表观遗传数据的信号分布(比如相对于TSS的距离)和信号强弱的组合设定。
举个例子:没有DNase-seq活性的区域,则不会分配调控元件信号;高DNase-seq活性的区域,缺乏H3K27ac,则会被判定为promoter-like elements;高DNase-seq活性的区域,具有H3K27ac活性,且是TSS-distal类型,则被判定为active distal enhancer-like elements。
三、Ensembl来源的enhancers
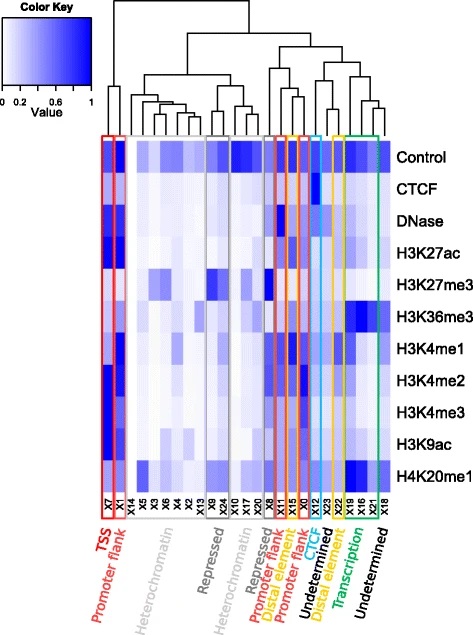
基于Segway segmentation states注释的方法(根据表观遗传信号的组合)
四、chromHMM定义的enhancers
这一部分可参照https://socrates1100.github.io/post/gong-neng-ji-yin-zu-zhu-shi/
和Ensembl来源的enhancers类似,是基于chromHMM segmentation states注释的方法(根据表观遗传信号的组合)
五、super enhancer (dbSuper)数据库来源的enhancers
基于H3K27ac ChIP-seq信号,该研究收集了86个组织细胞类型的58,283 super-enhancer regions。
References:
- http://bioinfo.vanderbilt.edu/AE/HACER/index.html
- Jing Wang, Xizhen Dai, Lynne D Berry, Joy D Cogan, Qi Liu, Yu Shyr, HACER: an atlas of human active enhancers to interpret regulatory variants, Nucleic Acids Research, Volume 47, Issue D1, 08 January 2019, Pages D106–D112
- https://enhancer.lbl.gov/
- The ENCODE Project Consortium., Moore, J.E., Purcaro, M.J. et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583, 699–710 (2020).
- Zerbino, D.R., Wilder, S.P., Johnson, N. et al. The Ensembl Regulatory Build. Genome Biol 16, 56 (2015).
- Roadmap Epigenomics Consortium., Kundaje, A., Meuleman, W. et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015).
- Aziz Khan, Xuegong Zhang, dbSUPER: a database of super-enhancers in mouse and human genome, Nucleic Acids Research, Volume 44, Issue D1, 4 January 2016, Pages D164–D171